 to bring up a window with just the protein sequence in FASTA format.
to bring up a window with just the protein sequence in FASTA format. The coursework assignment for this unit involves the classification and analysis of Amino-acyl tRNA synthetases. There are many bioinformatics packages/software available that could be used to do this. The aim of this workshop is to illustrate one method that could be used for the assignment for those unfamiliar with bioinformatics software, but others are equally valid.
During this workshop we will collect all the amino acyl tRNA synthetase sequences, then look at the structure of one (ArgRS), you will need to investigate them all for the coursework. Following this workshop you should attempt the complete coursework assignment given on moodle.
1. Search a sequence database for Thermus thermophilus amino-acyl tRNA synthetase sequences.
To find a sequence by name, you need to search the annotation accompanying the sequences, this can be done at several different websites. For this workshop we'll use those at EBI. To access the EBI, click here : http://www.ebi.ac.uk/.
We need to find the sequence of every trna synthetase from a single strain of T. thermophilus. I'll illustrate this using alanine, type Alanyl-trna synthetase AND thermus thermophilus into the search box and then search. Note that for some sequences you may need to use the equivalent Alanine trna ligase AND thermus thermophilus.
Results come up in several categories such as Genomes & Metagenomes, Nucleotide sequences, Protein sequences, etc.
Within the Protein sequences section, the T. thermophilus AlaRS sequences in the UniProtKB database are shown. These have been sequenced from different strains of Thermus. For the coursework you should try to choose a set of aaRS sequences all from the same strain. (You may need to click on the View all results for Protein Sequences link at the bottom of the Protein sequences section to display all the sequences).
Clicking on the link for the aaRS from your chosen strain will take you to the entry for that sequence in the UniProt database.
On the Uniprot page, scroll down to the Sequence section (can also be reached by clicking Sequence in the links bar on the left)
and click on  to bring up a window with just the protein sequence in FASTA format.
to bring up a window with just the protein sequence in FASTA format.
Copy this sequence, including the complete header line, into a wordpad, or notepad document, (NOT Word). Save this document with a meaningful name such as Thermus_aaRS.fasta
Now you have the Alanine tRNA synthetase protein sequence. Repeat this for the other 19 aaRS (actually this will be 20 as phenylalanyl trna synthetase has 2 subunits, making 21 sequences in total), putting all the protein sequences into the same file and save it in text format.
Make sure that each time you search for another aaRS you choose one from the same organism. For instance when I searched for PheRS the first two Protein sequences links were of unspecified strain and I had to look at those lower down the list to find the strain I'd been using.
When you have the complete set for T. thermophilus continue with the workshop below.
2. Alignment of the T. thermophilus aaRS sequences.
i) Organising the bacterial sequences into distinct classes using sequence alignment.
This part is similar
to the sequence alignment workshop script.
We will use T-Coffee to align the sequences. This is accessible from the ebi home page via the Services, Proteins links,
or here . Put your protein sequences into the box and click Submit . The T-Coffee results window contains the sequence alignment, and various tabs at the top. Click on the Phylogenetic
Tree tab. (If your browser crashes at this point, (Microsoft browsers used to have this issue) or doesn't show a tree, copy the whole link into another browser of your choice, e.g. Chrome or Firefox, then continue). This isn't a true phylogenetic tree, for that we'd need the DNA sequences, but a phylogram or cladogram, a visual representation of the similarity between the sequences. The sequences can be seen to cluster together into two rough groups. Compare these with the Classes of aaRS
introduced by Jean van den Elsen in his lectures. Note although you may obtain a visually-different tree from your neighbour, sequences can appear in a different order after alignment, but as long as the sequences cluster together within the same connection (i.e. the subclasses are together (not necessarily in the same order)), then the trees are identical.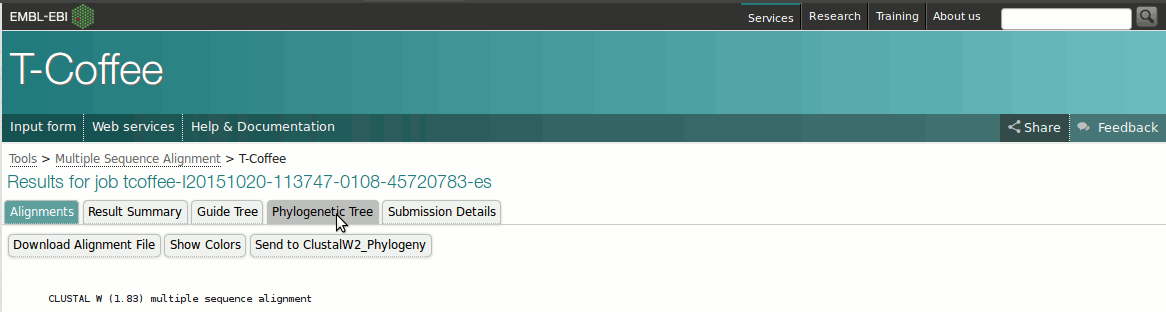
You might find it easier to analyse the tree if you change the labels drawn on the branches. This is done by altering what appears after the > symbol on the top line of each of the sequences and re-running T-coffee. Do not delete the rest of the header, you might need it later.
ii) Separate alignment of the two aaRS classes.
In order to make the 2 sequence alignments available on moodle, I separated the aaRS sequences into two groups based on the classes defined in the literature (and in Dr van den Elsen's lectures), and used T-Coffee to align these groups aswell. At this stage the proteins are gathered together into groups of shared similarity, so their alignment becomes more meaningful and it is possible to look for conservation of amino acids and structural domains within the alignment. We will not have time to do that in this workshop, but analysis of the two annotated alignments (which I have provided for you) is part of the coursework assessment.
3. Classifying the structural domains in the T. thermophilus Class 1 and Class II aaRS.
Comparing proteins structures visually can be quite challenging, however various classifications of structure have been devised. We will use the CATH classification, which you can read more about here (but don't need to do that now).
In this workshop we will illustrate how to analyse aaRS structures using one example; ArgRS, Class I.
If the structure of a protein has been elucidated it will be stored in the Protein Data Bank (PDB), the European version of which is available from the EBI.
Return to the EBI home page and click on Services on the top bar, then search for PDBe. On the results page, click on the PDBe  link. Here search for arginyl-tRNA synthetase and Thermus thermophilus. (note, it has to be arginyl, not arginine)
link. Here search for arginyl-tRNA synthetase and Thermus thermophilus. (note, it has to be arginyl, not arginine)
One structure is listed, given the PDB code 1iq0. Clicking on the link for 1iq0 brings up the PDBe page for the structure where you can find articles describing the structure solution, and links to other databases.
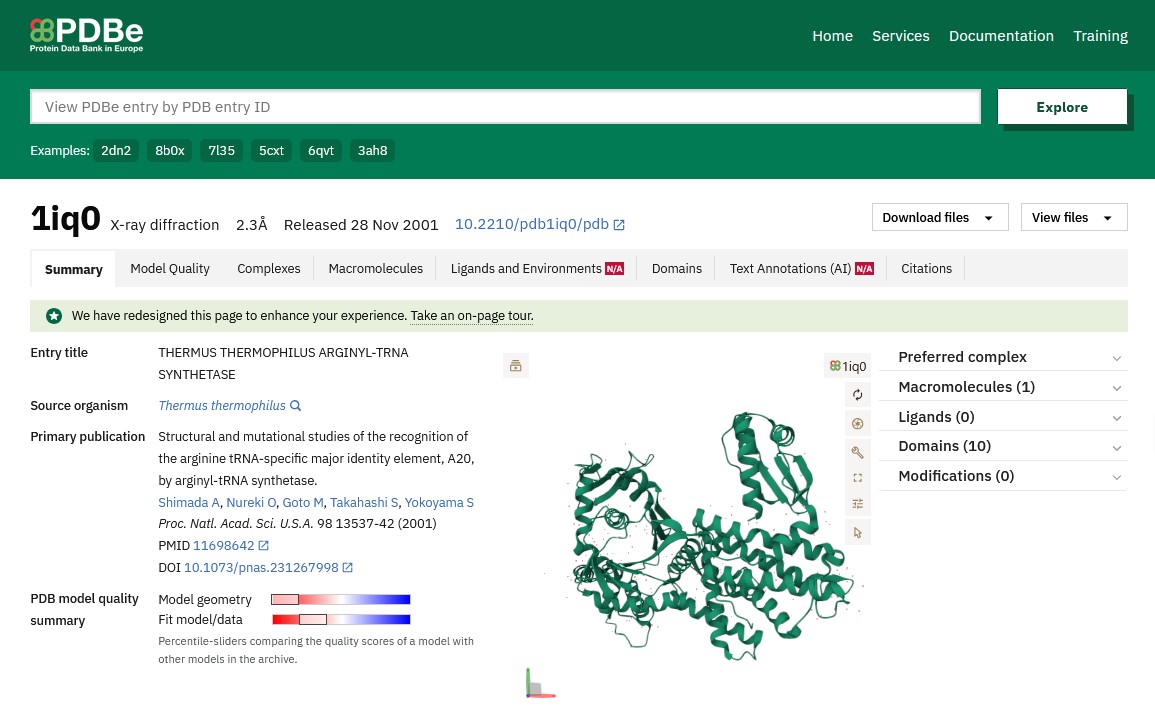
Looking at the schematic pictures of the structure at the top right of the page you can see that ArgRS has 1 protein chain in the structure, this is coloured green in the picture and is seen from 3 different directions but these pictures are not very informative about its function.
To find out more about the structure and the structural domains that it contains, click on Structure analysis on the right-hand bar, or further down this page. Some browsers crash trying to do this, if yours does, try a different browser.
On this new page it shows that the ArgRS is a monomer. Further down the page are links to the protein in a number of databases, including Pfam, which classifies proteins into functional families, and CATH: which classifies by structure. At the bottom of the page is a schematic diagram summarising what is known about the structure of ArgRS.
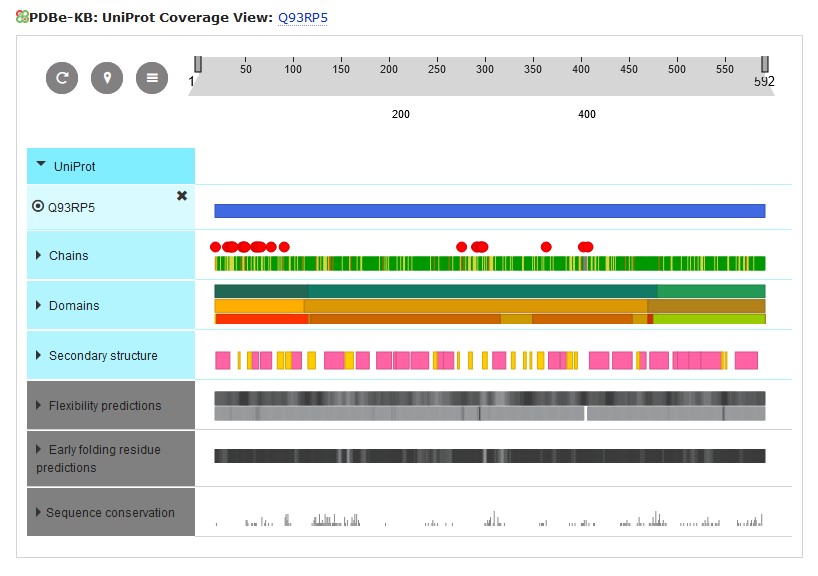
If you click on the Domains header, this section expands to show the domains making up the ArgRS. Hovering your mouse over each of the domains will show its code in that database.
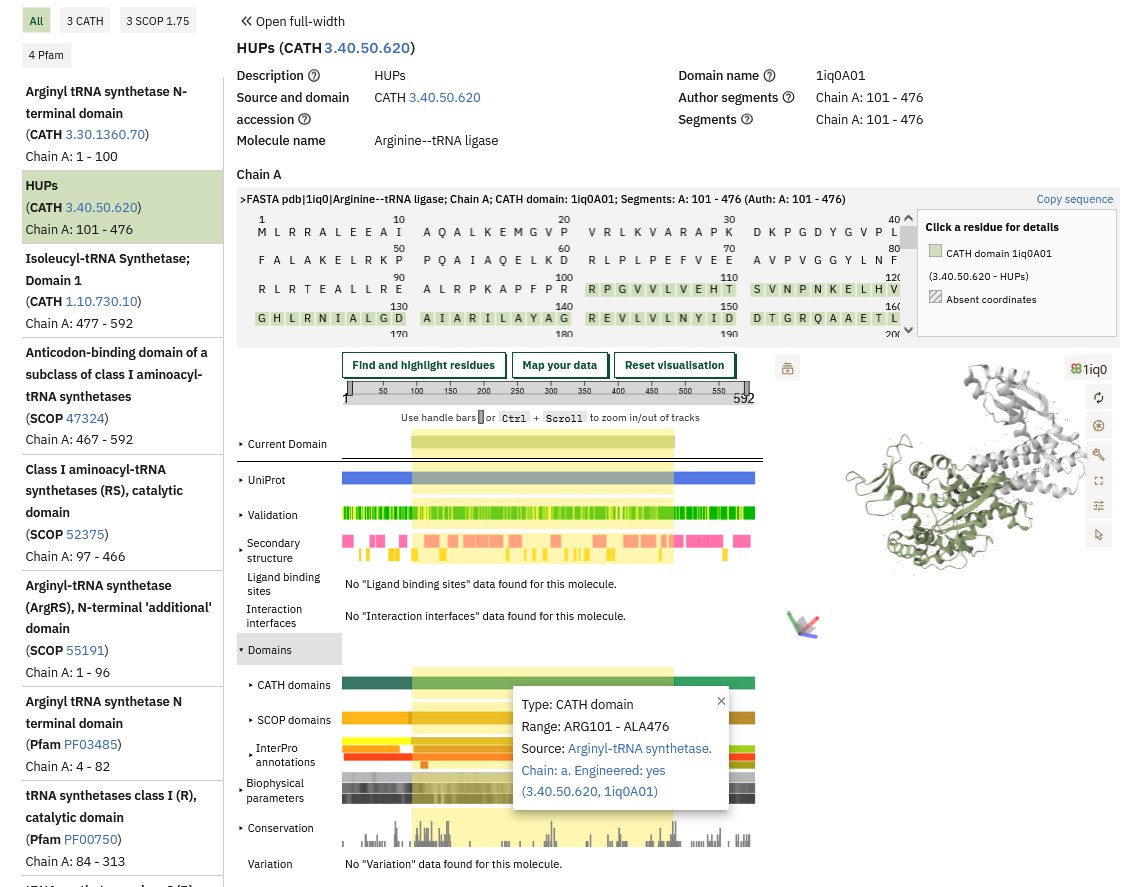
The Pfam database shows this is a tRNA synthetase (which we already knew). In the CATH line you can see that the ArgRS molecule contains 3 structural domains. Hovering over the CATH line will give the CATH numerical codes for these domains (the names given to the domains as listed above the diagram are not always relevant to the particular protein being investigated) and start and end of these domains within the ArgRS sequence, which will be useful in identifying these domains in the annotated Class I sequence alignment.
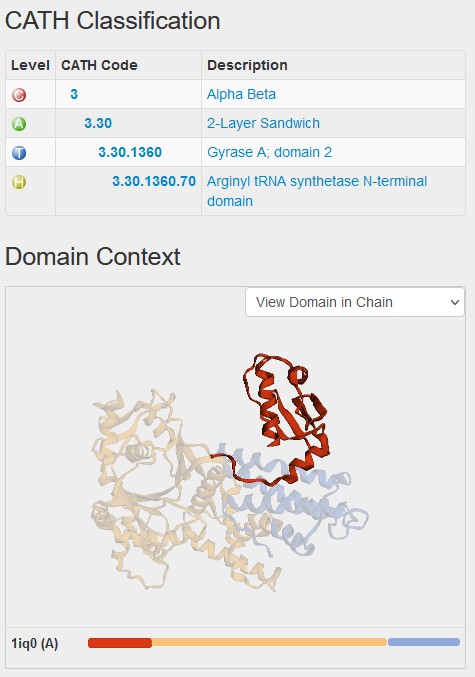
How this CATH classification relates to the structure can be seen in the CATH database by clicking on the link in the pop-up window above. This brings up the domain page in CATH database and shows you that it forms a small part of the structure, with two other domains coloured in yellow and blue. The classification of the first domain starts with a 3 because it is mixed alpha and beta.
5. Investigating domain function.
Finding out about domain function will mainly entail a literature search, combined with information from lectures. However, for some domains you can find functional information on the PDBe website. Returning to the top of the 1iq0 page in PDBe, click on the Function and Biology option on the right-hand menu.
At the bottom of this page, the CATH domains for the structure are listed (although these are not always listed in the same order as they fold in the sequence). Each has a schematic showing where it is in the structure. Next to these are the SCOP annotations, also with a schematic, and SCOP often includes the function of the domain.
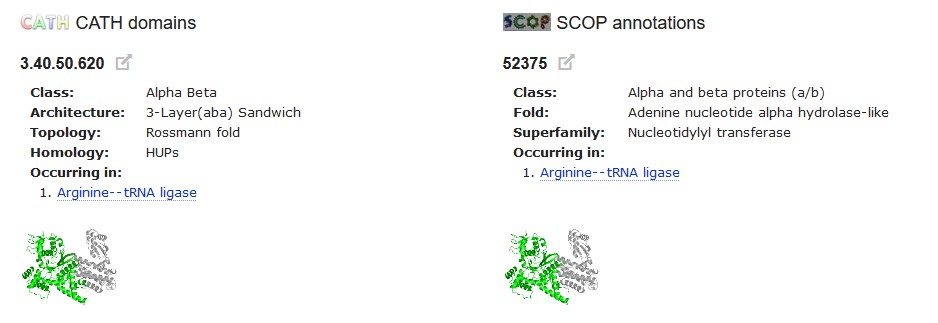
This shows us that the 3.40.50.620 domain has the 'Nucleotidylyl transferase' function, i.e. that it is the domain that joins the amino acid to its tRNA. Another of the domains' function is also identified here. The annotation of the third domain isn't so informative and you may need to read the paper on ArgRS to find out what its function is.
Having looked at the Function and Biology page, if you go back to the Structure Analysis page, you will see that SCOP functions can also be interrogated in the summary image at the bottom of the page.
Not all aaRS structures have this level of detail in their entries on PDBe, but it would be worth looking at all the Class I aaRS to see what can be obtained from PDBe before moving to other databases. Unfortunately not all the Thermus aaRS structures have been solved, so you may need to find the protein with the most similar sequence whose structure has been solved. Annoyingly PDBe has a sequence search limit of 1000 amino acids, which several of the aaRS exceed, so you need to use other resources.
Another 'secondary' structural database that may help to identify domains is PDBSUM. Entering the PDB codes of the structures you have identified, or the sequences you saved in the first part (entered without the header line), or even an annotation search with e.g. arginyl-trna synthetase, will bring up a PDBSUM page for the appropriate T. thermophilus aaRS structure, or the most similar. For example, try searching with the Uniprot code for the sequence you obtained for TrpRS. If you get no results, put the sequence in instead.
A PDBSUM page gives an overview of the reaction catalysed and the structure as well as links to many databases.
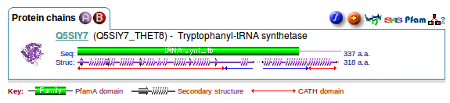 In the Protein chains section of the page you see a summary of the structural information, with two chains indicated by the labelled purple (A) and red (B) circles, a green bar linking to the Pfam database showing that this protein has tRNA synthetase activity, and the red and blue arrows underneath giving links to the CATH database. Again hovering over these gives you the codes for each domain.
In the Protein chains section of the page you see a summary of the structural information, with two chains indicated by the labelled purple (A) and red (B) circles, a green bar linking to the Pfam database showing that this protein has tRNA synthetase activity, and the red and blue arrows underneath giving links to the CATH database. Again hovering over these gives you the codes for each domain.
Clicking on an  link or on the Protein tab at the top takes you to
a page with more details, including an indication of the catalytic residues and the CATH classifications of both TrpRS domains. Check that the CATH domains identified within TrpRS agree with those you have already obtained and note any catalytic amino acids. The positions and type of catalytic amino acids are conserved within each aaRS Class.
link or on the Protein tab at the top takes you to
a page with more details, including an indication of the catalytic residues and the CATH classifications of both TrpRS domains. Check that the CATH domains identified within TrpRS agree with those you have already obtained and note any catalytic amino acids. The positions and type of catalytic amino acids are conserved within each aaRS Class.
On this Protein page the sequence is coloured by CATH domain, highlighting their position in the sequence, which should match their position in the Class I alignment, enabling you to start to match the CATH codes to the key of that alignment.
6. Now on to the Coursework.
Having identified the sequences and domains present in the structures of all the bacterial aaRS, (Class II structural data are collected for you and available from moodle) this would be your starting point to read the literature to find the missing domain functions and to answer the questions given in the coursework.